Now that we did some repeat modeling and classification, we can see if the breakpoints of any of the simulated inversions fall into these identified repeat regions.
library(dplyr)
library(ggplot2)
library(ggh4x)
library(tidyr)Output
Read in the repeat data¶
repeats <- read.table("repeat.regions", header = T)
repeats <- arrange(repeats[order(repeats$contig), ], contig, position_start)
# overwrite the file with it being sorted by chrom + start position
#write.table(repeats, file = "repeat.regions", quote = F, row.names = F)
nrow(repeats)
head(repeats, 15)Read in inversion inventory¶
We’ll to read in the inventory of all inversion positions and pull out the unique inversions. I really should have committed to either comma or tab separated tables.
inversions <- read.csv("../assess_sv_leviathan/visor/linkedread.sv.assessment", header = T)
inversions <- unique(inversions[, c(1,6,9,10)])
rownames(inversions) <- NULL
inversions$is_repeat <- as.character(NA)
inversions$which_breakpoints <- 0
inversions$is_repeat_start <- as.character(NA)
inversions$is_repeat_end <- as.character(NA)
head(inversions)Assess which inversions are in or adjacent to repeat regions¶
To do the matchy-matchy, similar to what we did to assess inversion calling. It will need some modifications to accommodate the slightly different use case. Primarily, adding a 800bp buffer to the repeat breakpoints to catch inversion breakpoints that might be within an insert-size of the repeat breakpoint. The which_breakpoints column codes either 0 for “none”, 1 for first-only, 2 for second-only, and 3 for both.
for(i in 1:nrow(inversions)){
.row <- inversions[i,]
# first breakpoint
query <- which(
repeats$contig == .row$contig &
(repeats$position_start - 800 <= .row$position_start & repeats$position_end + 800 >= .row$position_start)
)
if(length(query) > 0) {
inversions$is_repeat_start[i] <- paste(repeats$class[query], collapse = ";")
inversions$which_breakpoints[i] <- inversions$which_breakpoints[i] + 1
}
# second breakpoint
query <- which(
repeats$contig == .row$contig &
(repeats$position_start - 800 <= .row$position_end) & (repeats$position_end + 800 >= .row$position_end)
)
if(length(query) > 0) {
inversions$is_repeat_end[i] <- paste(repeats$class[query], collapse = ";")
inversions$which_breakpoints[i] <- inversions$which_breakpoints[i] + 2
}
}
inversions$is_repeat <- !is.na(inversions$is_repeat_start) | !is.na(inversions$is_repeat_end)
are_repeats <- inversions[inversions$is_repeat, ]
are_repeatsRead in inversion results¶
Let’s find which inversions had the most false negatives.
false_neg <- read.csv("../assess_sv_leviathan/visor/linkedread.sv.assessment", header = T) %>%
group_by(contig, position_start, position_end, size) %>%
mutate(perc = round(sum(assessment == "false negative")/length(depth) * 100,1)) %>%
filter(assessment == "false negative") %>%
summarize(n = length(depth), perc = unique(perc)) %>%
ungroup() %>%
arrange(desc(n))
false_neg$is_repeat <- FALSE
false_neg$which_breakpoint <- as.integer(0)
head(false_neg)`summarise()` has grouped output by 'contig', 'position_start', 'position_end'.
You can override using the `.groups` argument.
We can now combine the inversions-in-repeats with the inversions-not-detected dataframe to see if there’s a correlation of hard-to-call inversions being associated with repeat regions.
for(i in 1:nrow(false_neg)){
.row <- false_neg[i,]
query <- which(
are_repeats$contig == .row$contig &
are_repeats$position_start == .row$position_start
)
if(length(query) > 0) {
false_neg$is_repeat[i] <- TRUE
false_neg$which_breakpoint[i] <- are_repeats$which_breakpoint[query]
}
}
false_neg$platform <- "linkedread"
head(arrange(false_neg, desc(is_repeat)))For comparison sake, we need to do this with the long-read inversion detections as well. We’ll read it in and extract only the relevant columns and only rows where assessment is false negative.
long_inv <- read.csv("../longread_workflow/longread.sv.assessment", header = T)
false_neg_long <- group_by(long_inv[,c(2:5, 8,10)], contig, position_start, position_end, size, platform) %>%
mutate(perc = round(sum(assessment == "false negative")/length(assessment) * 100,1)) %>%
filter(assessment == "false negative") %>%
summarize(n = length(assessment), perc = unique(perc)) %>%
ungroup() %>%
arrange(desc(n))
false_neg_long$is_repeat <- FALSE
false_neg_long$which_breakpoint <- as.integer(0)
head(false_neg_long, 20)`summarise()` has grouped output by 'contig', 'position_start', 'position_end',
'size'. You can override using the `.groups` argument.
Now do the same matching between inversions and repeat regions
for(i in 1:nrow(false_neg_long)){
.row <- false_neg_long[i,]
query <- which(
are_repeats$contig == .row$contig &
are_repeats$position_start == .row$position_start
)
if(length(query) > 0) {
false_neg_long$is_repeat[i] <- TRUE
false_neg_long$which_breakpoint[i] <- are_repeats$which_breakpoint[query]
}
}
head(arrange(false_neg_long, desc(is_repeat)))Let’s combine the two dataframes so we can start plotting them to meaningfully represent these data. This will also include making platform, size, and contig ordered factors.
false_neg_all <- rbind(false_neg, false_neg_long) %>% arrange(desc(n), desc(platform), desc(size))
false_neg_all$contig <- factor(false_neg_all$contig, levels = c("2L", "2R","3L","3R"), ordered = T)
false_neg_all$size <- factor(false_neg_all$size, levels = c("small", "medium", "large", "xl"))
false_neg_all$platform <- factor(false_neg_all$platform, levels = c("linkedread","ontshort","ontlong","pacbio"))
head(false_neg_all)It may be worth adding a column of unique identifiers for the inversions for plotting purposes.
false_neg_all <- group_by(false_neg_all, is_repeat,position_start) %>%
mutate(inv_id = cur_group_id())
false_neg_all$seq_id <- 1:nrow(false_neg_all)
head(false_neg_all, 15)Visualization¶
Source
tech_colors <- c(
"ontshort" = "#9b6981",
"ontlong" = "#682c37",
"pacbio" = "#f6955e" ,
"linkedread" = "#a8cdec"
)
size_colors <- c(
"#c0c6cf",
"#a2aab5",
"#6e7684",
"#54546c"
)Output
options(repr.plot.width = 22, repr.plot.height = 7)
ggplot(false_neg_all, aes(x = seq_id, y = 0, yend = perc, shape = size, color = platform)) +
geom_point(size = 4, aes(y = perc)) +
geom_segment(aes(linetype = is_repeat), linewidth = 1.1) +
theme_light() +
scale_shape_manual(values = c(19,15,18,17)) +
scale_color_manual(values = tech_colors) +
coord_cartesian(xlim = c(-2, 195), ylim = c(0, 101),expand = F) +
labs(
title = "False Negatives in Repeat Regions",
color = "Technology",
shape = "Inversion Size",
linetype = "In Repeat Region"
) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank()
) +
xlab("Inversion") +
ylab("Percent False Negative Detection Across Samples")
options(repr.plot.width = 20, repr.plot.height = 10)
.strip <- strip_themed(background_y = elem_list_rect(fill = tech_colors))
false_neg_all %>% group_by(platform) %>%
mutate(mx = max(perc)) %>%
arrange(desc(mx), .by_group = T) %>%
mutate(id = 1:length(perc)) %>%
ggplot(aes(x = id, y = 0, yend = perc, shape = size, color = size)) +
geom_point(size = 6, aes(y = perc)) +
geom_segment(aes(linetype = is_repeat), linewidth = 2) +
theme_light() +
scale_shape_manual(values = c(19,15,18,17)) +
scale_color_manual(values = size_colors) +
labs(
title = "False Negatives in Repeat Regions",
color = "Inversion Size",
shape = "Inversion Size",
linetype = "In Repeat Region"
) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major.x=element_blank(),
panel.grid.minor.x=element_blank()
) +
xlab("Inversion") +
ylab("Percent False Negative Detection Across Samples") +
facet_grid2(rows = vars(platform), strip = .strip)
options(repr.plot.width = 20, repr.plot.height = 10)
.strip <- strip_themed(background_y = elem_list_rect(fill = size_colors))
false_neg_all %>% group_by(size) %>%
mutate(mx = max(perc)) %>%
arrange(desc(mx), .by_group = T) %>%
mutate(id = 1:length(perc)) %>%
ggplot(aes(x = id, y = 0, yend = perc, shape = platform, color = platform)) +
geom_point(size = 5, aes(y = perc)) +
geom_segment(aes(linetype = is_repeat), linewidth = 1.5) +
theme_light() +
scale_shape_manual(values = c(19,15,18,17)) +
scale_color_manual(values = tech_colors) +
labs(
title = "False Negatives in Repeat Regions",
color = "Technology",
shape = "Technology",
linetype = "In Repeat Region"
) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major.x=element_blank(),
panel.grid.minor.x=element_blank()
) +
xlab("Inversion") +
ylab("Percent False Negative Detection Across Samples") +
facet_grid2(rows = vars(size), scales = "free_x", independent = "x", strip = .strip)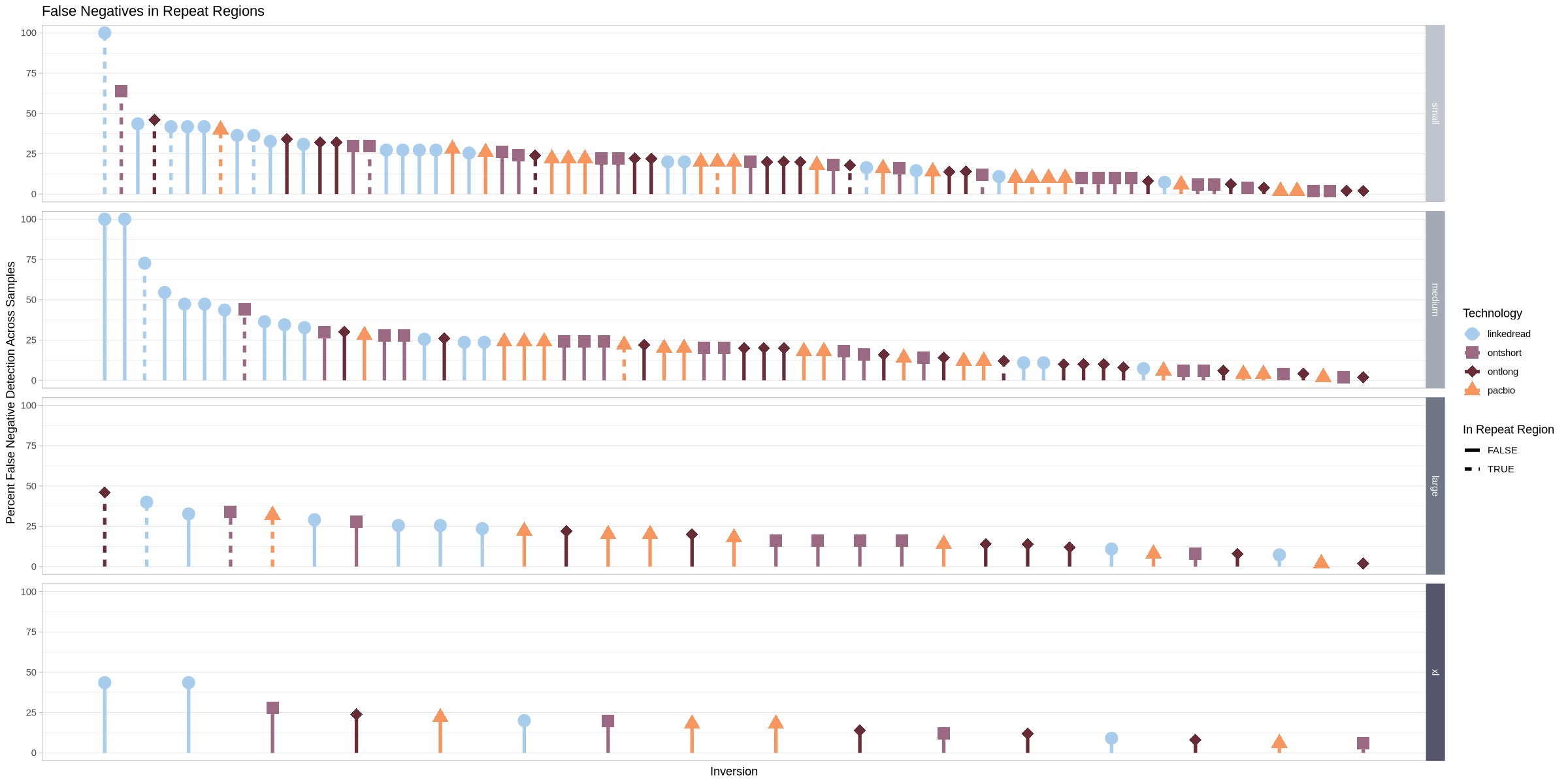
freq_colors <- c(
"#a4c0d8ff",
"#a86cafff"
)
options(repr.plot.width = 20, repr.plot.height = 8)
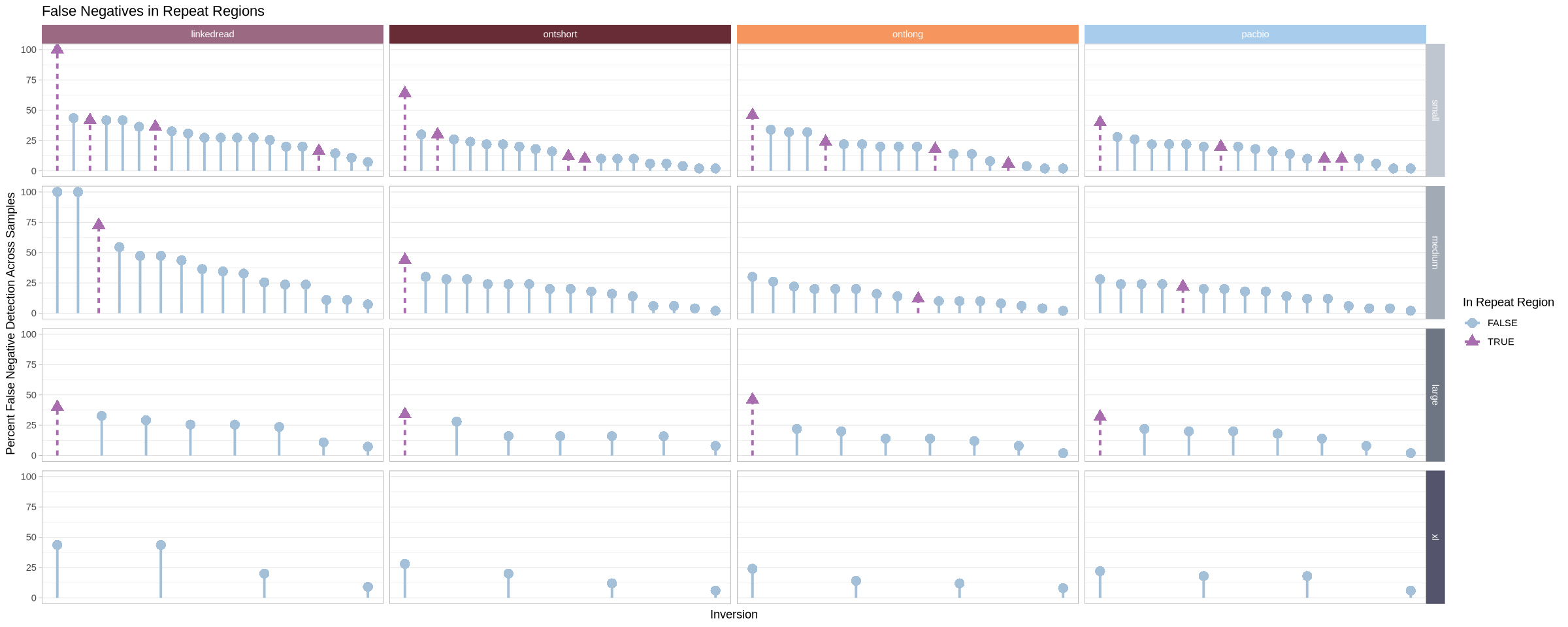
.strip <- strip_themed(background_x = elem_list_rect(fill = tech_colors), background_y = elem_list_rect(fill = size_colors))
false_neg_all %>% group_by(platform, size) %>%
mutate(mx = max(perc)) %>%
arrange(desc(mx), .by_group = T) %>%
mutate(id = 1:length(perc)) %>%
ggplot(aes(x = id, y = 0, yend = perc, shape = is_repeat, color = is_repeat)) +
geom_point(size = 4, aes(y = perc)) +
geom_segment(aes(linetype = is_repeat), linewidth = 1.1) +
theme_light() +
#scale_shape_manual(values = c(19,15,18,17)) +
scale_color_manual(values = freq_colors) +
labs(
title = "False Negatives in Repeat Regions",
color = "In Repeat Region",
shape = "In Repeat Region",
linetype = "In Repeat Region"
) +
theme(
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major.x=element_blank(),
panel.grid.minor.x=element_blank()
) +
xlab("Inversion") +
ylab("Percent False Negative Detection Across Samples") +
facet_grid2(rows = vars(size), cols = vars(platform), scales = "free_x", independent = "x", strip = .strip)